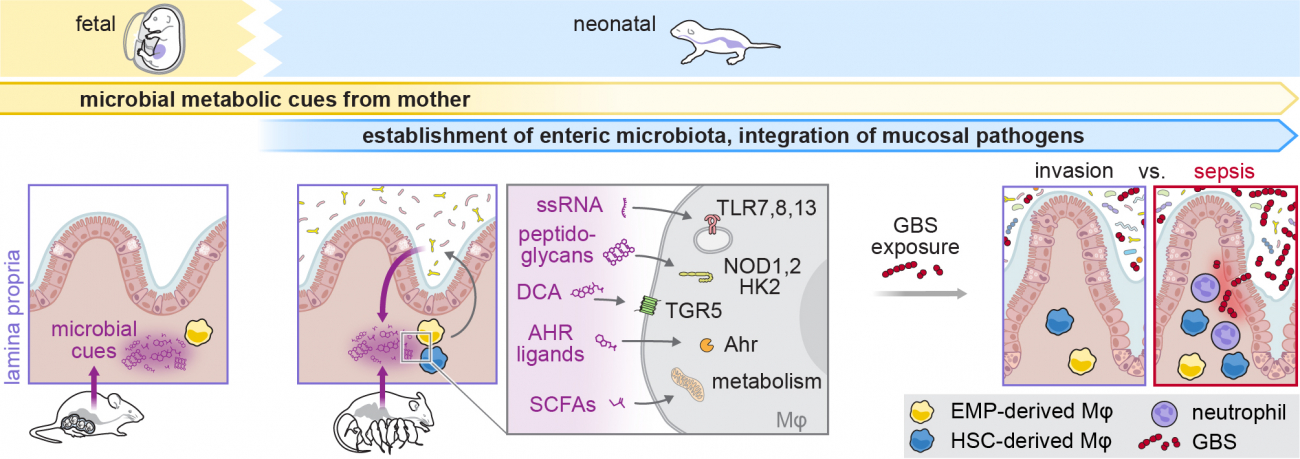
Perinatal crosstalk of microbiota and intestinal macrophages in the control of mucosal pathogens
Our project explores the co-development of the neonatal microbiota and intestinal Mφ. We focus on metabolic programming and epigenetic imprinting in the integration of potentially highly pathogenic mucosal colonizers, in particular streptococci, into the host-adapted microbiota. We examine molecular and cellular mechanisms underlying co-evolution of a stable intestinal host-microbe interface.
Team
Publications
Prospective Whole-Genome Sequencing to Identify Bacterial Transmission and Its Modifiers in Neonates |
Trained immunity in skin infections: Macrophages and beyond2025 ELife
|
Sensory neurons shape local macrophage identity via TGF-β signalingKolter J, Döring CL, Sarout S, Baasch S, Steele L, Alsumati R, Lucena Silva GV, Aníbal Silva CE, Paiva IM, Mansoori Moghadam Z, Gres V, Lohrmann F, Aktories P, Buchegger T, Bijnen M, Doumard L, Voisin B, Le Foll C, Lachmann N, Greter M, Kierdorf K, Haniffa M, Cunha TM, Flacher V, Henneke P 2025 Immunity
|
Immunopathogenic and clinical implications of advanced tissue analysis in non-tuberculous mycobacterial infections in childrenMaximilian Seidl, Elham Bavafaye Haghighi, Anne Kathrin Lösslein, Markus Hufnagel, Florens Lohrmann, Christian Schneider, Daniela S Kohlfürst, Werner Zenz, Gregor Gorkiewicz, Cornelia Feiterna-Sperling, Renate Krüger, Peter Bronsert, Christina Neppl, Kim Zoe Sommer, Verena Stehl, Melanie Boerries, Martin Kuntz, Philipp Henneke 2025 Frontiers in Immunology
|
The infant microbiota hopscotches between community states toward maturation—longitudinal stool parameters and microbiota development in a cohort of European toddlersIntze E, Schaubeck M, Pourjam M, Neuhaus K, Lagkouvardos I, Hitch TCA, Clavel T. 2025 ISME Communications
|
Reactive oxygen species regulate early development of the intestinal macrophage-microbiome interfaceMansoori Moghadam Z, Zhao B, Raynaud C, Strohmeier V, Neuber J, Lösslein AK, Qureshi S, Gres V, Ziegelbauer T, Baasch S, Schell C, Warnatz K, Inohara N, Nunez G, Clavel T, Rosshart SP, Kolter J, Henneke P. 2025 Blood
|
Salmonella infection accelerates postnatal maturation of the intestinal epitheliumStefan Schlößer, Anna-Lena Ullrich, Nastaran Fazel Modares, Matthias A. Schmitz, Johannes Schöneich, Kaiyi Zhang, Isabel Richter, Laura Robrahn, Sarah Schraven, James S. Nagai, Sven-Bastiaan Haange, Susan A. V. Jennings, Thomas Clavel, Ulrike Rolle-Kampczyk, Fabian Kiessling, Ivan G. Costa, Vanesa Muncan, Urska Repnik, Martin von Bergen, Aline Dupont, and Mathias W. Hornef 2024 PNAS
|
M cell maturation and cDC activation determine the onset of adaptive immune priming in the neonatal Peyer’s patchTorow N, Li R, Hitch TCA, Mingels C, Al Bounny S, van Best N, Stange EL, Simons B, Maié T, Rüttger L, Gubbi NMKP, Abbott DA, Benabid A, Gadermayr M, Runge S, Treichel N, Merhof D, Rosshart SP, Jehmlich N, Hand TW, von Bergen M, Heymann F, Pabst O, Clavel T, Tacke F, Lelouard H, Costa IG, Hornef MW 2023 Immunity
|